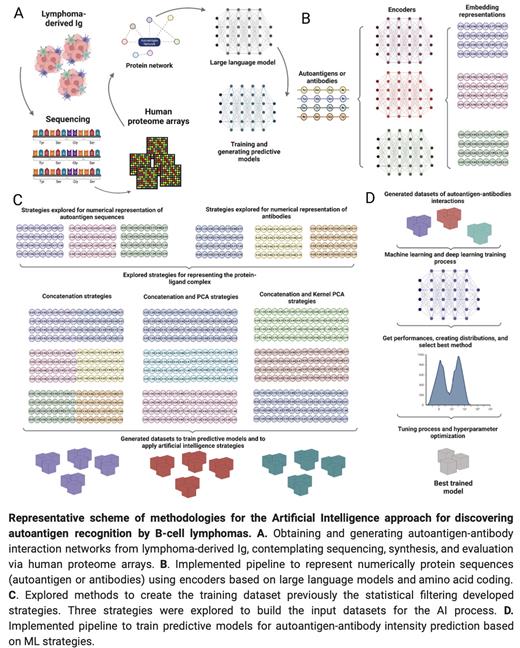
Artificial Intelligence approach for the discovery of autoantigen recognition by B-cell lymphomas
Introduction
With a common origin from mature B-cells, B-cell lymphomas express a unique clonal surface immunoglobulin (Ig) that may transmit survival signals autonomously or following binding to cognate antigens. An increasing number of potential autoantigen targets is described for several mature B-cell lymphomas, however, the discovery of novel targets represents a very complex and expensive task.
Thanks to the development of artificial intelligence (AI) tools such as AlphaFold and advances in natural language processing methods such as Large Language Models, the study of proteins has benefited, making it possible to accelerate studies of interactions between proteins. In this context, we explored the training of AI models to predict autoantigen recognition of lymphoma-derived Ig from linear protein information.
Methods
First, 45 lymphoma-derived Ig were sequenced, synthesized as recombinant proteins, and probed onto human proteome arrays generating 370,000 antigens-antibodies interactions. Next, statistical methods were designed and implemented to reduce noise and filter the autoantigen-antibody interactions to be processed. Subsequently, sequence-based methods were explored to implement and validate predictive models of autoantigen-antibody interaction intensity. Lastly, sequence similarity network strategies were analyzed to identify preference relationships between antibodies and autoantigens.
Results and Discussion
From the 370,000 interactions, designed filters allowed to reduce the noise, generating a total of 270,000 valid interactions, which were used to train predictive models. Large language model methods, amino acid coding strategies via physicochemical properties, and spatial transformation techniques through Fourier transforms were explored as methods for the numerical representation of autoantigen and antibody sequences. Concatenation strategies and linear and non-linear combinations were explored to represent the autoantigen-antibody interaction complexes. More than 1000 predictive models were explored. The best performances were obtained by applying the pre-trained bepler and esm1b models, concatenation strategies, and using Random Forest algorithms as a training strategy. In addition, cross-validation methods with k-fold (k=10) were applied to prevent overfitting. The best results achieve a performance of 0.9 of Pearsons' coefficient and an MSE of 0.08. Alternatively, training strategies based on deep learning architectures such as CNN or GCN were used, although they presented similar results to those achieved by the selected method. Finally, the model was validated using molecular dynamics techniques, studying the affinity of interactions in a selected random sample, with a correlation between the predicted affinity results and the affinity of the interaction complexes measured by molecular dynamics.
Conclusions
The large language model methods explored for the training of predictive models have been combined with sequence similarity network methods for constructing autoantigen and antibody interaction networks. Interaction networks helped to identify patterns of antibody recognition preferences. In future work, the incorporation of unsupervised learning algorithms, enrichment analysis, and simulation of interactions via the generated predictive model is proposed to build an efficient pattern detection strategy to facilitate the discovery of autoantigen interaction of lymphoma derived Ig.
Disclosures
No relevant conflicts of interest to declare.